Evolutionary Hyper Parameter optimisation for Deep Learning
Art of finding the right parameters for a DNN. Hyper parameter optmisation is an art in Deep Learning. Evolutionary algorithms are well suited for search an optimisation tasks with massive search space, such as this. In this post I make use of FSharp to optimse a DNN written in Keras with Tensorflow backend.
Disclosure: This article was first published in Medium (https://medium.com/ixmx/evolutionary-hyper-parameter-optimisation-with-fsharp-keras-tensorflow-62bf441e19cd) by myself.
I have been away from FSharp for a while. Advent calendar event organised by Sergey Tihon seemed like a good time to get back in to the scene. Coming back to .Net and FSharp is always a challenge. Platforms, Frameworks, tooling all had changed, new ways of doing old things and all for the better.
I’ve decided to try out the .Net Core as it seems to be the future. My previous FSharp installation on the Mac was running Mono so this time I’ve created a linux docker container and installed .Net Core in it. Problem with .Net is the amount of choice you have. So many platforms to choose with pros and cons. This is in stark contrast to another language — Golang which I use heavily these days. Getting up and running on Golang is pretty straightforward and quick.
The whole purpose of this is to try out various evolutionary algorithm experiments I do. These algorithms have good properties that are better implemented in a functional language. Normally an EA will include a Genetic Algorithm (GA), which is a search and optimisation algorithm with directed random search.
Genetic Algorithm
A GA is suited for problems with massive search space, as such I’ve used it to optimise hyper parameters of a Deep Neural Network (DNN). Usually DNN implementation would be done in Python with Keras as the DNN library and with TensorFlow backend performing neural graph computations. This setup is widely used but I’m not comfortable relying on Python to do any complicated code than a simple script. In Machine Learning, where the results could take hours or days, runtime error at the end of an experiment which could’ve been detected at compile time ruining your week is not the risk I would take. Fsharp’s strict static typing comes handy here. Also its terseness without sacrificing readability shines as well.
Deep Learning
Typical Deep Learning experiment would be a some sort of a classifier, regardless of the data. A successfully trained model depends on selecting the proper hyper parameters, which is mostly an art. Typical Convolutional Neural Network (CNN) contains a number of filters with kernels of various sizes.
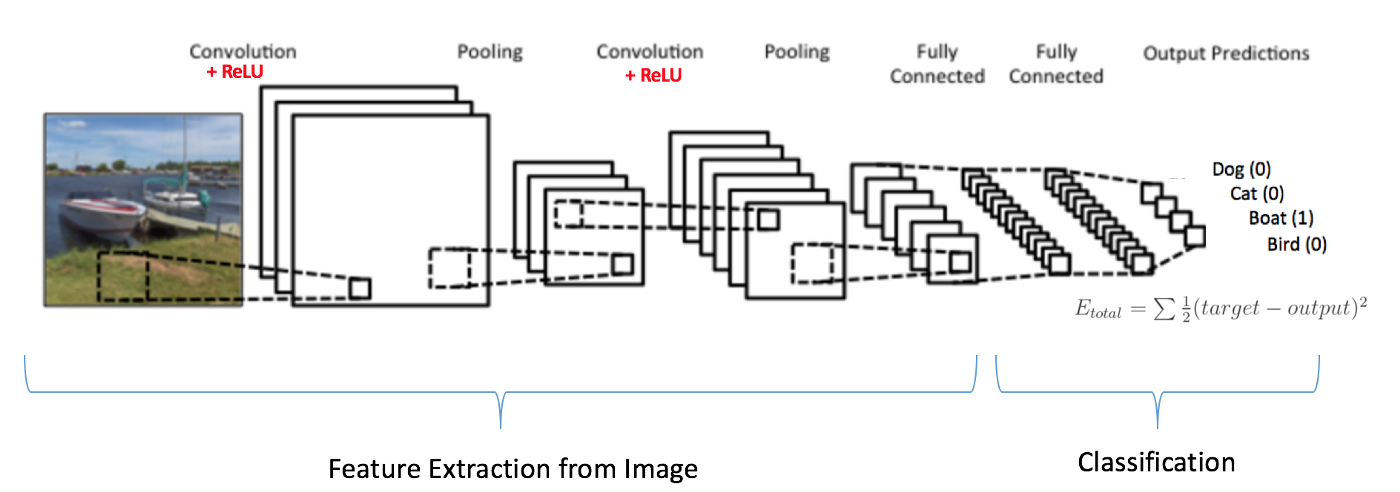 Structure of a CNN. Image Credit: https://www.kdnuggets.com/2016/11/intuitive-explanation-convolutional-neural-networks.html/3
Structure of a CNN. Image Credit: https://www.kdnuggets.com/2016/11/intuitive-explanation-convolutional-neural-networks.html/3
These convolutional layers are pooled into other layers as a way of subsampling. Each pooling stage will have a pooling window which again have to be decided. Then comes the Dense Layers size of which again has to be decided. Drop out is another hyper parameter that needs tuning, along with batch size and epoch count. Drop out refers to number of connections to drop from a layer, batch size refers to the size of input data used before backpropagation updates the network weights. Epoch count refers to the number of training iterations to perform before stopping the experiment.
Depending on these parameters used, you may get a model which has good error rate, accuracy and/or length of training. We can consider each experiment as an action performed by an agent. Each agent will have the above mentioned hyper parameters as its properties. Each agent will then compete in a tournament to create a model which has some winning characteristics. We evolve the agents in each generation selecting the most viable. Therefore an agent can be modelled as below.
An DNN hyper parameters are then mapped into a Chromosome (GA terminology). A Chromosome is an array of floating point values (ranging from 0.0 to 1.0), that would be subjected to the GA operators. Here they would be randomly mutated and spliced with other successful Chromosomes, over generations. Typical GA would have the following steps:
In each generation, the population of Agents is evaluated.
During the evaluation, each Chromosome is mapped back to actual DNN hyper parameters and the DNN training script (Python) would be executed with these as command line arguments. Inside the Python script these values are used to construct the DNN architecture.
The Chromosome values have to be mapped to actual ranges of each of the Agent properties before its evaluation. For example, the following illustrates this process. The number of DNN hyper parameters determines the length of a Chromosome.
The experiment_runner.sh is a simple bash file to run the Python script and filter out the unwanted bits from Keras output. It typically looks like below.
This yields an output similar to one below:
The output parsing code should extract the val_loss and/or val_acc values to formulate the performance of the experiment.
Once the agents are evaluated, they are assigned with a performance score, or a Fitness which will be used for evolving the next generation. The selection strategy we use here is called tournament selection where one half of the population is compared against the other half and the agent with lower fitness inherits a changed Chromosome. This process is shown in the following function.
Once the agents are evaluated, they are assigned with a performance score, or a Fitness which will be used for evolving the next generation. The selection strategy we use here is called tournament selection where one half of the population is compared against the other half and the agent with lower fitness inherits a changed Chromosome. This process is shown in the following function.
On the Python script, command line arguments are received as such:
With these variables a Tensorflow graph can be created using Keras layers as below:
Running these experiments take a while. When we have to run each experiment (agent) on each generation for a number of generations, we are looking at a few days (at least) of computation. This can be parallelised by evaluating the whole generation in one go. In order to do that, we need to use a cluster of cloud instances with two endpoints, one to execute the experiment with given hyper parameters and another to check for the progress and get the performance metric.